Modules¶
regressors.stats¶
-
regressors.stats.sse(clf, X, y)[source]¶ Calculate the standard squared error of the model.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
Returns: The standard squared error of the model.
Return type: float
-
regressors.stats.adj_r2_score(clf, X, y)[source]¶ Calculate the adjusted
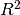 of the model.
of the model.Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
Returns: The adjusted
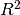 of the model.
of the model.Return type: float
-
regressors.stats.coef_se(clf, X, y)[source]¶ Calculate standard error for beta coefficients.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
Returns: An array of standard errors for the beta coefficients.
Return type: numpy.ndarray
-
regressors.stats.coef_tval(clf, X, y)[source]¶ Calculate t-statistic for beta coefficients.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
Returns: An array of t-statistic values.
Return type: numpy.ndarray
-
regressors.stats.coef_pval(clf, X, y)[source]¶ Calculate p-values for beta coefficients.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
Returns: An array of p-values.
Return type: numpy.ndarray
-
regressors.stats.f_stat(clf, X, y)[source]¶ Calculate summary F-statistic for beta coefficients.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
Returns: The F-statistic value.
Return type: float
-
regressors.stats.residuals(clf, X, y, r_type=u'standardized')[source]¶ Calculate residuals or standardized residuals.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
- r_type (str) –
Type of residuals to return: ‘raw’, ‘standardized’, ‘studentized’. Defaults to ‘standardized’.
- ‘raw’ will return the raw residuals.
- ‘standardized’ will return the standardized residuals, also known as internally studentized residuals, which is calculated as the residuals divided by the square root of MSE (or the STD of the residuals).
- ‘studentized’ will return the externally studentized residuals, which is calculated as the raw residuals divided by sqrt(LOO-MSE * (1 - leverage_score)).
Returns: An array of residuals.
Return type: numpy.ndarray
-
regressors.stats.summary(clf, X, y, xlabels=None)[source]¶ Output summary statistics for a fitted regression model.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
- xlabels (list, tuple) – The labels for the predictors.
regressors.plots¶
-
regressors.plots.plot_residuals(clf, X, y, r_type=u'standardized', figsize=(10, 8))[source]¶ Plot residuals of a linear model.
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
- r_type (str) –
Type of residuals to return: ‘raw’, ‘standardized’, ‘studentized’. Defaults to ‘standardized’.
- ‘raw’ will return the raw residuals.
- ‘standardized’ will return the standardized residuals, also known as internally studentized residuals, which is calculated as the residuals divided by the square root of MSE (or the STD of the residuals).
- ‘studentized’ will return the externally studentized residuals, which is calculated as the raw residuals divided by sqrt(LOO-MSE * (1 - leverage_score)).
- figsize (tuple) – A tuple indicating the size of the plot to be created, with format (x-axis, y-axis). Defaults to (10, 8).
Returns: The Figure instance.
Return type: matplotlib.figure.Figure
-
regressors.plots.plot_qq(clf, X, y, figsize=(7, 7))[source]¶ Generate a Q-Q plot (a.k.a. normal quantile plot).
Parameters: - clf (sklearn.linear_model) – A scikit-learn linear model classifier with a predict() method.
- X (numpy.ndarray) – Training data used to fit the classifier.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
- figsize (tuple) – A tuple indicating the size of the plot to be created, with format (x-axis, y-axis). Defaults to (7, 7).
Returns: The Figure instance.
Return type: matplotlib.figure.Figure
-
regressors.plots.plot_pca_pairs(clf_pca, x_train, y=None, n_components=3, diag=u'kde', cmap=None, figsize=(10, 10))[source]¶ Create pairwise plots of principal components from x data.
Colors the components according to the y values.
Parameters: - clf_pca (sklearn.decomposition.PCA) – A fitted scikit-learn PCA model.
- x_train (numpy.ndarray) – Training data used to fit clf_pca, either scaled or un-scaled, depending on how clf_pca was fit.
- y (numpy.ndarray) – Target training values, of shape = [n_samples].
- n_components (int) – Desired number of principal components to plot. Defaults to 3.
- diag (str) –
Type of plot to display on the diagonals. Default is ‘kde’.
- ‘kde’: density curves
- ‘hist’: histograms
- cmap (str) – A string representation of a Seaborn color map. See available maps: https://stanford.edu/~mwaskom/software/seaborn/tutorial/color_palettes.
- figsize (tuple) – A tuple indicating the size of the plot to be created, with format (x-axis, y-axis). Defaults to (10, 10).
Returns: The Figure instance.
Return type: matplotlib.figure.Figure
-
regressors.plots.plot_scree(clf_pca, xlim=[-1, 10], ylim=[-0.1, 1.0], required_var=0.9, figsize=(10, 5))[source]¶ Create side-by-side scree plots for analyzing variance of principal components from PCA.
Parameters: - clf_pca (sklearn.decomposition.PCA) – A fitted scikit-learn PCA model.
- xlim (list) – X-axis range. If required_var is supplied, the maximum x-axis value will automatically be set so that the required variance line is visible on the plot. Defaults to [-1, 10].
- ylim (list) – Y-axis range. Defaults to [-0.1, 1.0].
- required_var (float, int, None) – A value of variance to distinguish on the scree plot. Set to None to not include on the plot. Defaults to 0.90.
- figsize (tuple) – A tuple indicating the size of the plot to be created, with format (x-axis, y-axis). Defaults to (10, 5).
Returns: The Figure instance.
Return type: matplotlib.figure.Figure
regressors.regressors¶
-
class
regressors.regressors.PCR(n_components=None, regression_type=u'ols', alpha=1.0, l1_ratio=0.5, n_jobs=1)[source]¶ Principal components regression model.
This model solves a regression model after standard scaling the X data and performing PCA to reduce the dimensionality of X. This class simply creates a pipeline that utilizes:
- sklearn.preprocessing.StandardScaler
- sklearn.decomposition.PCA
- a supported sklearn.linear_model
Attributes of the class mimic what is provided by scikit-learn’s PCA and linear model classes. Additional attributes specifically relevant to PCR are also provided, such as
PCR.beta_coef_.Parameters: - n_components (int, float, None, str) –
Number of components to keep when performing PCA. If n_components is not set all components are kept:
n_components == min(n_samples, n_features)
If n_components == ‘mle’, Minka’s MLE is used to guess the dimension. If
0 < n_components < 1, selects the number of components such that the amount of variance that needs to be explained is greater than the percentage specified by n_components. - regression_type (str) – The type of regression classifier to use. Must be one of ‘ols’, ‘lasso’, ‘ridge’, or ‘elasticnet’.
- n_jobs (int (optional)) – The number of jobs to use for the computation. If
n_jobs=-1, all CPUs are used. This will only increase speed of computation for n_targets > 1 and sufficiently large problems. - alpha (float (optional)) – Used when regression_type is ‘lasso’, ‘ridge’, or ‘elasticnet’.
Represents the constant that multiplies the penalty terms. Setting
alpha=0is equivalent to ordinary least square and it is advised in that case to instead useregression_type='ols'. See the scikit-learn documentation for the chosen regression model for more information in this parameter. - l1_ratio (float (optional)) – Used when regression_type is ‘elasticnet’. The ElasticNet mixing
parameter, with
0 <= l1_ratio <= 1. Forl1_ratio = 0the penalty is an L2 penalty.For l1_ratio = 1it is an L1 penalty. For0 < l1_ratio < 1, the penalty is a combination of L1 and L2.
-
scaler¶ sklearn.preprocessing.StandardScaler, None
The StandardScaler object used to center the X data and scale to unit variance. Must have
fit()andtransform()methods. Can be overridden prior to fitting to use a different scaler:pcr = PCR() # Change StandardScaler options pcr.scaler = StandardScaler(with_mean=False, with_std=True) pcr.fit(X, y)
The scaler can also be removed prior to fitting (to not scale X during fitting or predictions) with pcr.scaler = None.
-
prcomp¶ sklearn.decomposition.PCA
The PCA object use to perform PCA. This can also be accessed in the same way as the scaler.
-
regression¶ sklearn.linear_model
The linear model object used to perform regression. Must have
fit()andpredict()methods. This defaults to OLS using scikit-learn’s LinearRegression classifier, but can be overridden either using the regression_type parameter when instantiating the class, or by replacing the regression model with a different on prior to fitting:pcr = PCR(regression_type='ols') # Examine the current regression model print(pcr.regression) LinearRegression(copy_X=True, fit_intercept=True, n_jobs=1, normalize=False) # Use Lasso regression with cross-validation instead of OLS pcr.regression = linear_model.LassoCV(n_alphas=200) print(pcr.regression) LassoCV(alphas=None, copy_X=True, cv=None, eps=0.001, fit_intercept=True, max_iter=1000, n_alphas=200, n_jobs=1, normalize=False, positive=False, precompute='auto', random_state=None, selection='cyclic', tol=0.0001, verbose=False) pcr.fit(X, y)
-
beta_coef_¶ Returns: Beta coefficients, corresponding to coefficients in the original space and dimension of X. These are calculated as 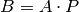 , where
, where 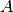 is a vector of the
coefficients obtained from regression on the principal components
and
is a vector of the
coefficients obtained from regression on the principal components
and 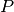 is the matrix of loadings from PCA.
is the matrix of loadings from PCA.Return type: numpy.ndarray
-
fit(X, y)[source]¶ Fit the PCR model.
Parameters: - X (numpy.ndarray) – Training data.
- y (numpy.ndarray) – Target values.
Returns: An instance of self.
Return type: regression.PCR
-
intercept_¶ Returns: The intercept for the regression model, both in PCA-space and in the original X-space. Return type: float